I have 500+ text files to read into Exploratory, and they are pipe-delimited:
This blog post gives an elegant solution for reading in multiple CSV files:
How to Read Multiple Excel or CSV Files Together
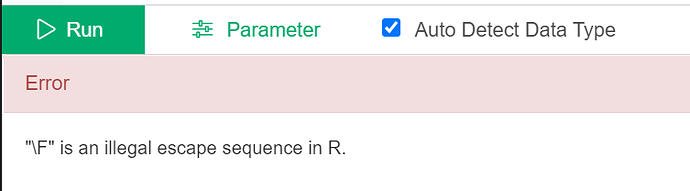
But when I try it, here is the result:
How can I specify that the delimiter is “|”?
sugiaki
February 2, 2021, 1:00am
#2
Hi @Stephanie_Brady
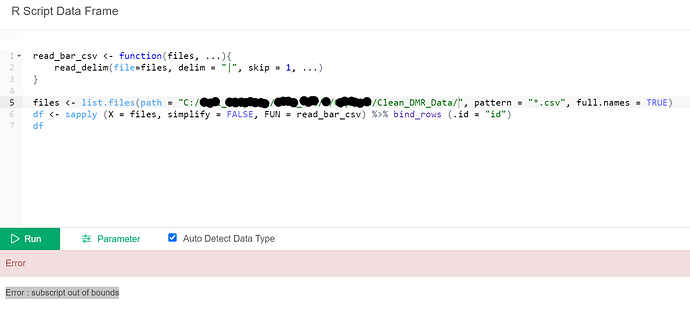
How about the following solution? Please adjust the file path to your environment and run the following script in the R script data frame of Exploratory.
It may take a bit of time to load 500+ text files at once, so another solution may be needed…I hope this helps.
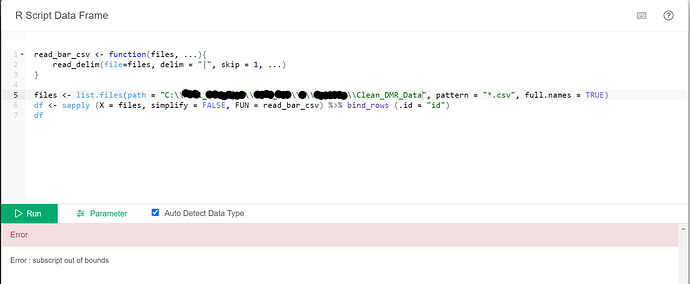
read_bar_csv <- function(files, ...){
read_delim(file = files, delim = "|", skip = 1, ...)
}
files <- list.files(path = "~/Desktop/multiple_files/", pattern = "*.csv", full.names = TRUE)
df <- sapply(X = files, simplify=FALSE, FUN = read_bar_csv) %>% bind_rows(.id = "id")
df
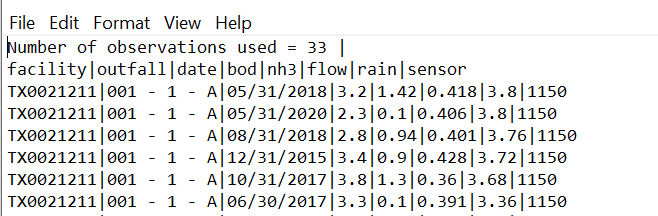
I have prepared three sample csv data with pipe-delimited in the format you specify, as shown below.
> cat ~/Desktop/multiple_files/test001.csv
Number of observations used = 33|
facility|outfall|date|bod|nh3|flow|rain|sensor
TX00211|001 - 1 - A|05/31/2018|3.2|1.42|0.418|1|1150
TX00211|001 - 1 - A|05/31/2018|3.3|1.43|0.419|2|1151
TX00211|001 - 1 - A|05/31/2018|3.4|1.44|0.420|3|1152
TX00211|001 - 1 - A|05/31/2018|3.5|1.45|0.421|4|1153
~
> cat ~/Desktop/multiple_files/test002.csv
Number of observations used = 35|
facility|outfall|date|bod|nh3|flow|rain|sensor
TX00212|002 - 1 - A|05/31/2018|3.1|1.52|1.418|1|2150
TX00212|002 - 1 - A|06/31/2018|3.3|1.63|3.419|2|2151
TX00212|002 - 1 - A|07/31/2018|3.2|1.74|2.420|3|2152
TX00212|002 - 1 - A|08/31/2018|3.5|1.85|1.421|4|2153
~
> cat ~/Desktop/multiple_files/test003.csv
Number of observations used = 33|
facility|outfall|date|bod|nh3|flow|rain|sensor
TX00213|003 - 1 - A|05/31/2018|3.2|1.42|0.418|1|1150
TX00213|003 - 1 - A|05/31/2018|3.3|1.43|0.419|2|1151
TX00213|003 - 1 - A|05/31/2018|3.4|1.44|0.420|3|1152
TX00213|003 - 1 - A|05/31/2018|3.5|1.45|0.421|4|1153
2 Likes
Hi Stephanie,\\ instead of /)
path = "C:\\Test\\Folder"
@sugiaki thank you so much for the detailed post!
I made a few different attempts relating to replacing the file path and keep getting this error:
Any ideas what I can try next?
@hide_kojima1
Thank you, I am on a Windows machine.
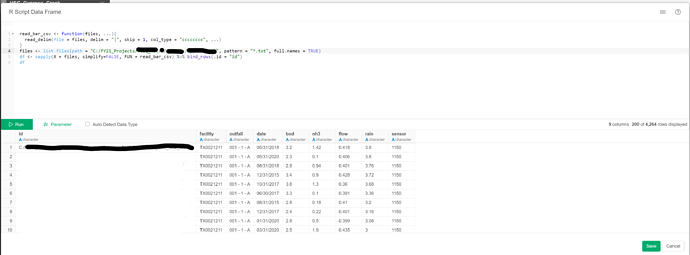
My apologies, I actually have “.txt” files. Changing the extension (pattern = “*.txt”) worked great!
read_bar_csv <- function(files, ...){
read_delim(file = files, delim = "|", skip = 1, ...)
}
files <- list.files(path = "C:/.../.../Test/Folder", pattern = "*.txt", full.names = TRUE)
df <- sapply(X = files, simplify=FALSE, FUN = read_bar_csv) %>% bind_rows(.id = "id")
df
1 Like
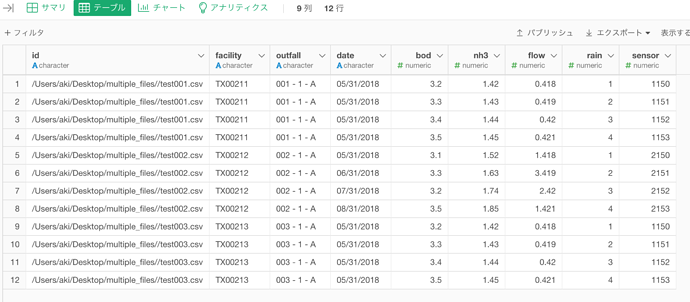
One more update. There was some variation in my input files, and I had trouble importing all of them at once due to issues with how the data type was detected. I clicked the “Auto Detect Data Type” option off and it didn’t force the data types to character.
I looked into the read_delim function and learned how to set the col_type , as shown below:
@hide_kojima1 indicated that
if you uncheck the “Auto Detect Data Type”, since you use read_delim in your R script, it honors the the data type detected by the read_delim.
Thank you both for your help!
1 Like